
Welcome to my personal website
You will find here information about me, my research, and my side projects. Feel free to contact me if you have any questions.

Welcome to my personal website
You will find here information about me, my research, and my side projects. Feel free to contact me if you have any questions.


I'm a big data enthusiast!
I have a Ph.D. in biology and have studied microorganisms for many years. I love analyzing data, programming, and bioinformatics. Those are my primary domains, but I don't fear transitioning to another field that would interest me.
I have years of experience utilizing various microscopy and staining techniques. I also work in a molecular laboratory, but my biggest passion is analyzing data.
In my free time, I like cooking, repairing old electronics, studying math, and coding for fun. If you want to know more about me, check my CV or feel free to contact me.
As environmental molecular data are still increasing, many specialists can benefit from this data. They can assess the diversity of the group of their interest. One way to do it is to use the phylogenetic placement method - placing the environmental sequences (based on the phylogenetic signal) on the reference tree of already identified taxa. But reference trees are missing for many groups. One of the examples are ciliates. Therefore, we decided to fill this gap and design a global ciliate SSU-rRNA tree that may be used in future studies.
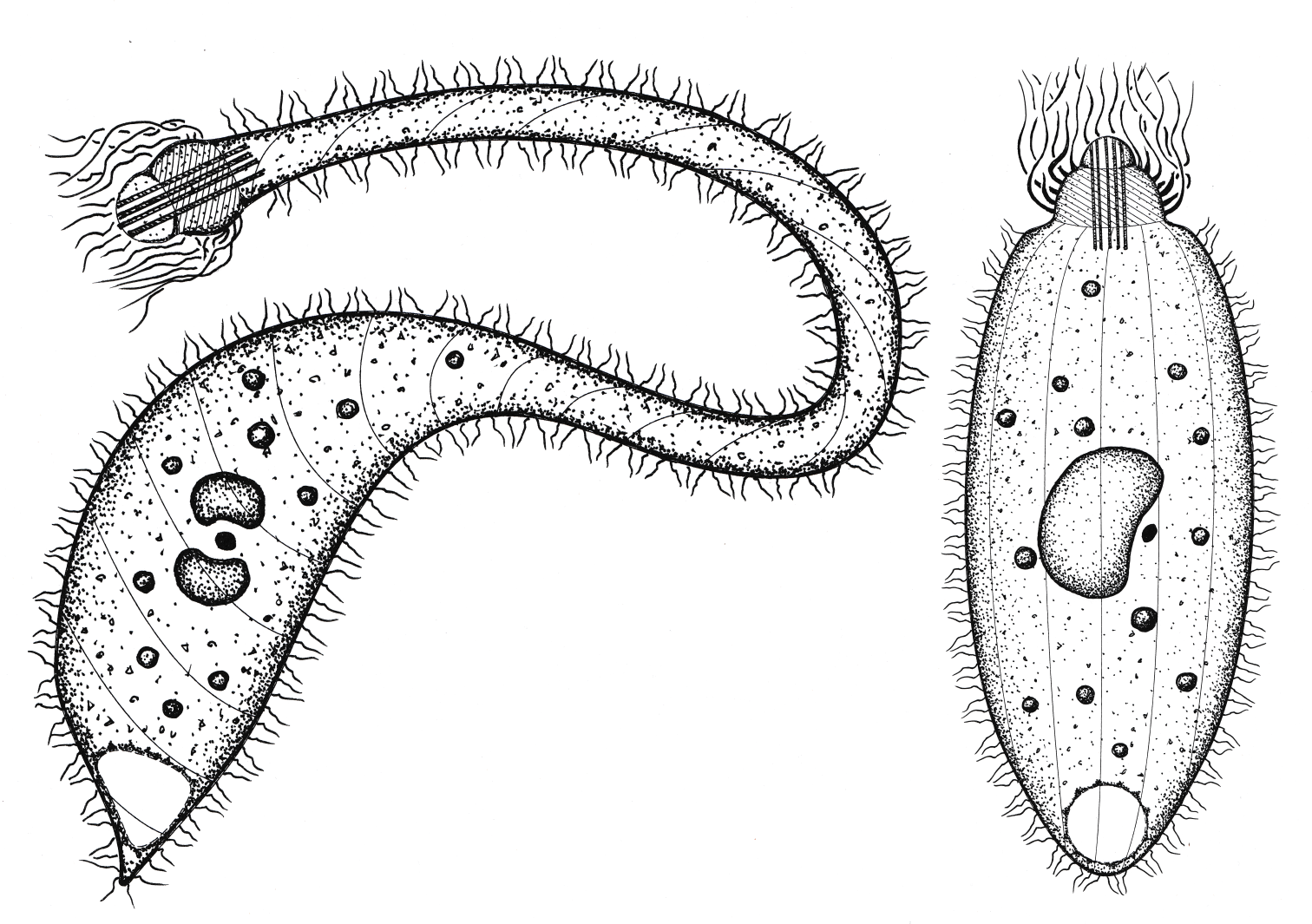
This study points out that the formal characterization of protists should include more than one type of evidence, for example, only DNA sequence data. We used a combination of molecular and morphological tools such as 18S rRNA sequences and various microscopy techniques.
The majority of ciliate phylogenetic studies today are based on ribosomal genes or currently rising genomic data. Yet many evolutionary questions are still unresolved, and we often do not know what barriers stand between successful and unsuccessful evolutionary reconstruction. To find out what can hinder the phylogenetic analyses, we focused only on one protein-coding gene: alpha-tubulin.
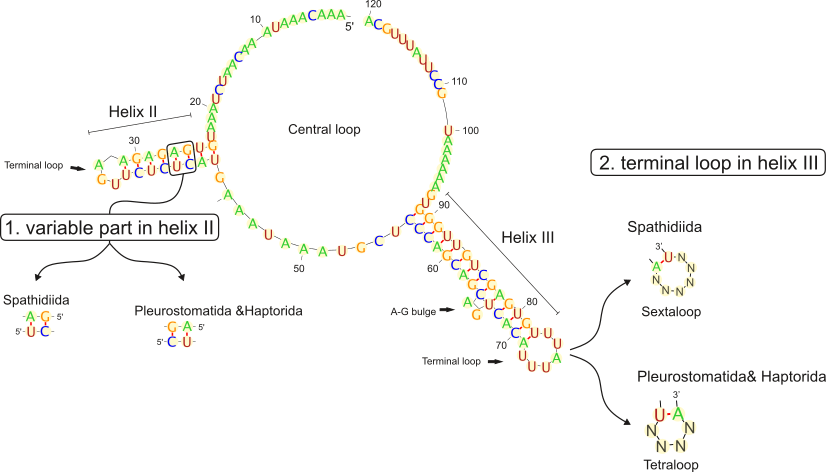
As we added molecular markers and increased the sampling for litostomatean ciliates, it was possible to analyze the evolutionary relationships among the four main free-living litostomatean lineages (Haptorida, Pleurostomatida, Rhynchostomatia, and Spathidiida) for the first time.
Recent detailed phylogenetic studies uncover the discrepancy between classical morphological and molecular classification of predatory ciliates from the order Spathidiida. We found out that several rapid radiations (prolific formation of new taxa) shaped their evolutionary history in the past.
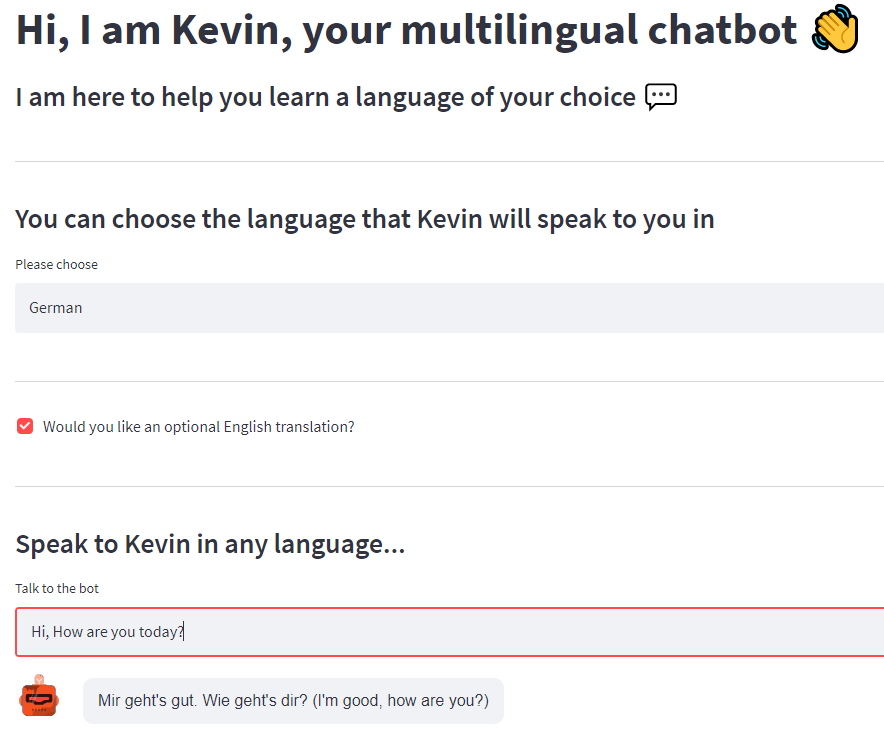
This project started during the project week at Le Wagon Bootcamp. Four of us - Nicolai Lewis, Ewa Rusinowska, Bhavika Srivastava, and me - wanted to build a conversational AI platform where you can practice a language of your choice anytime and for free. So, we built a multilingual deep learning chatbot named Kevin.
Kevin can:
Feel free to chat with Kevin. Have fun and sharpen your language skills anytime-anywhere and for free :)