Main Objective
This study points out that the formal characterization of protists should include more than one type of evidence, for example, only DNA sequence data. We used a combination of molecular and morphological tools such as 18S rRNA sequences and various microscopy techniques. Due to this integrative approach, we were able to test our hypotheses using independent data and make more robust analyses.
Morphological and molecular work
I collected and investigated samples during my research stay in Boise, Idaho, USA. There, I learned different morphological methods from my host Dr. William Bourland, one of the leading experts on ciliates, and Dr. Xiaotian Luo, a talented young scientist. We investigated various protists from freshwater, soil, and sand samples. The first step was a live observation using the light microscope and thorough documentation of all ciliates encountered. I found one relatively rare ciliate species called Phialina pupula. As this species is not well-documented, I decided to look closer at the population I found. To uncover all the intricate morphological details of such a tiny animal, we used silver (protargol) staining and scanning electron microscopy.
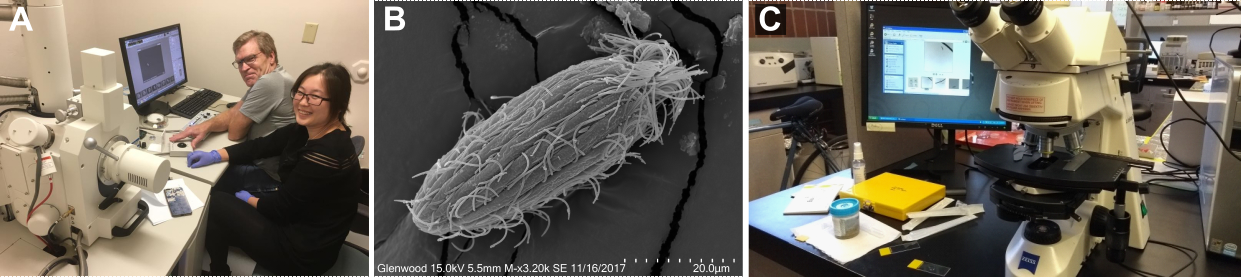
Figure 1. (A) Dr. William Bourland and Dr. Xiaotian Luo using scanning electron microscope, (B) Phialina from the scanning electron microscopy, (C) Investigation of all the samples under the light microscope.
I also extracted, amplified, and cloned the ribosomal 18S-ITS1-5.8S-ITS2 rDNA. After molecular cloning, the DNA was again amplified and purified. And then finally sent for the sequencing. After obtaining sequences, we made phylogenetic analyses to reconstruct the evolutionary history of the species and its related family members on the tree of life.
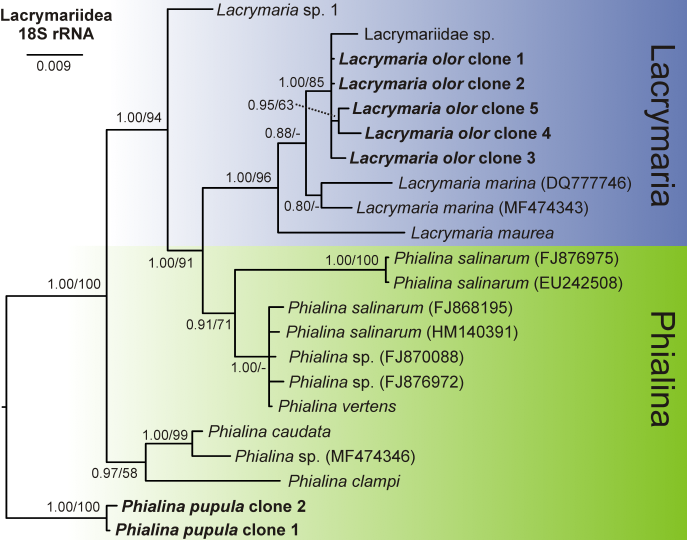
Figure 2. Phylogeny based on the 18S rRNA gene of 22 taxa from the family Lacrymariidae.
Conclusion
Morphological studies are time-consuming, and the diversity of microorganisms is immense. Therefore, molecular methods gradually became more and more popular. In particular, the introduction of next-generation sequencing that allows detecting many taxa in the environment. These molecular studies showed that the diversity of microorganisms is underestimated, and many taxa that were thought to be rare are probably just overlooked. As we still don't know what organisms are around us, the classical species (re-)descriptions are still essential for exploring Earth's diversity. Our study represents a modern take on such a species re-description. We provide morphological details (from live observation, staining, and scanning electron microscopy) as well as molecular data (from multiple molecular markers that showed to be phylogenetically informative).