Litostomatean Phylogeny
As we added molecular markers and increased the sampling for litostomatean ciliates, it was possible to analyze the evolutionary relationships among the four main free-living litostomatean lineages (Haptorida, Pleurostomatida, Rhynchostomatia, and Spathidiida) for the first time.
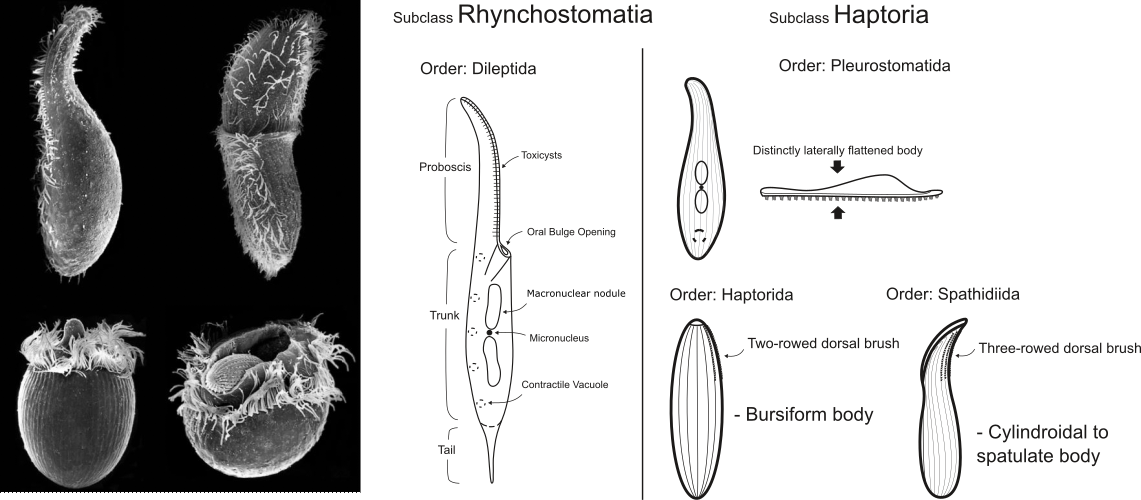
Figure 2. Overall morphology of the marvelous predatory ciliates from the class Litostomatea. You can see the main morphological differences between the main litostomatean lineages.
Analyzing the 18S rRNA gene and the ITS region sequences uncovered the following:
1) The presence of a primary bifurcation of the litostomateans into two main groups: the subclasses Rhynchostomatia and Haptoria. This primary branching was also supported by the secondary structure of the ITS2 molecule.
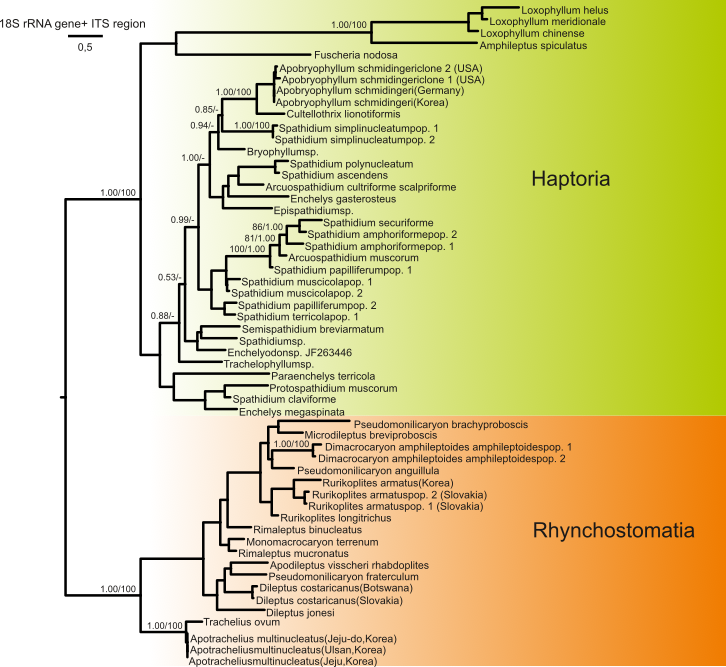
Figure 2. Phylogeny based on the 18S rRNA gene and the ITS1-5.8S-ITS2 region of 56 litostomatean taxa showing the primary phylogenetic bifurcation into Rhynchostomatia and Haptoria.
2) The presence of three lineages (orders) within the subclass Haptoria: Haptorida, Pleurostomatida, and Spathidiida. The Spathidiida lineage appeared as a separate branch in our analyses, whereas the Haptorida and Pleurostomatida lineages were sister lineages. Several shared morphological features also back this relationship: meridionally extending somatic kineties in the anterior region of the cell and reduced number of dorsal brush rows. Spathidiids, on the other hand, are characterized by anteriorly curved somatic kineties and a three-rowed dorsal brush. The secondary structure of the ITS2 molecule and the results of quartet mapping also favored a sister relationship between the Haptorida and Pleurostomatida.
3) An ancestrally unique motif in the secondary structure of the ITS2 molecule indicated monophyly of the order Spathidiida. This monophyly is also supported by anteriorly bent somatic kineties and a three-row dorsal brush.
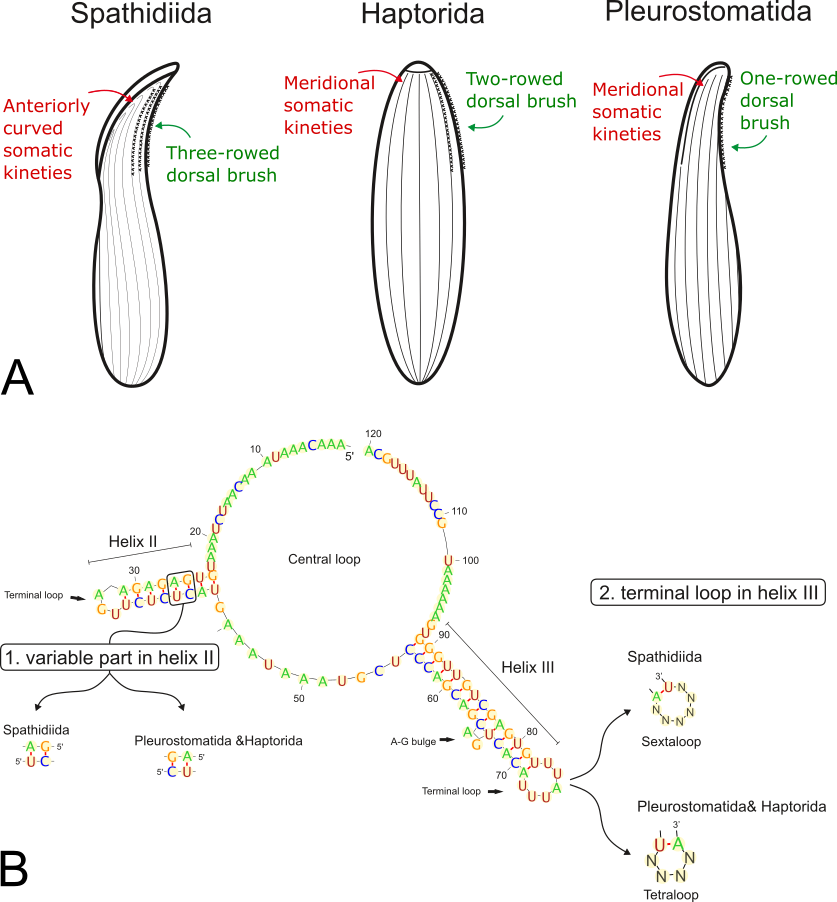
Figure 1. (A) Somatic apomorphies of the investigating haptorians orders, (B) Consensus secondary structureof the ITS2 molecule of free-living litostomateans and the molecular synapomorphies of its main lineages.
Conclusion
Utilization of the 18S rRNA gene along with the ITS region is revealed to be beneficial for phylogenetic inferences in ciliates, because concatenation of the comparatively conservative 18S rRNA gene sequences with the faster evolving ITS region sequences might lead to increasing statistical support in phylogenetic trees. Moreover, the conservative parts of helix II and III of the ITS2 molecule might serve as a good proxy for reconstruction of not only recent but also deep evolutionary events. And finally molecular phylogenetic analyses of the 18S rRNA gene and the ITS region harmonize well with assumptions based on the structural conservatism hypothesis, postulating that somatic ciliary structures are phylogenetically more informative than oral patterns.