Main Objective
In this study, we focused on three major problems people often face during phylogenetic or phylogenomic analyses based on the protein-coding genes such as alpha-tubulin. 1) Insufficient phylogenetic signal, 2) presence of selection pressure, and 3) molecular homoplasies.
Insufficient phylogenetic signal
All our alpha-tubulin trees had limited phylogenetic resolution, with only one distinct and statistically well-supported clade. In contrast, the other taxa formed independent lineages or were in a basal polytomy. This indicates that only a few phylogenetically informative positions exist in both nucleotide and amino acid sequences.
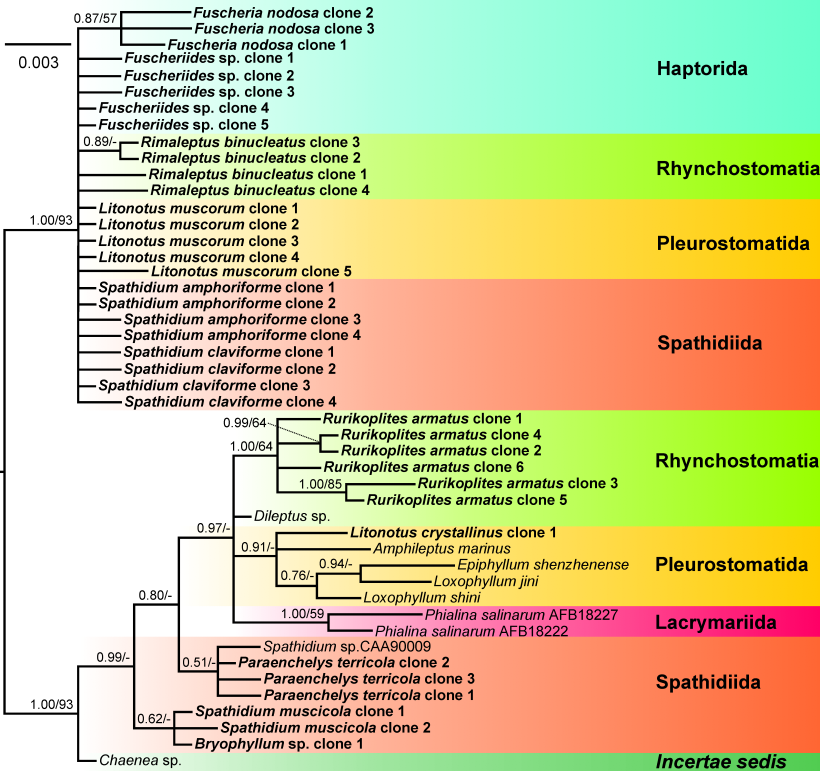
Figure 1. Amino acid tree showing low phylogenetic resolution.
Presence of selection pressure
When we focused on individual amino acid positions, the SIMMAP analysis revealed 30 positions under positive selection and up to 297 positions under negative selection out of the total 357 positions. These analyses indicate that litostomatean alpha-tubulin is strongly affected by selection.
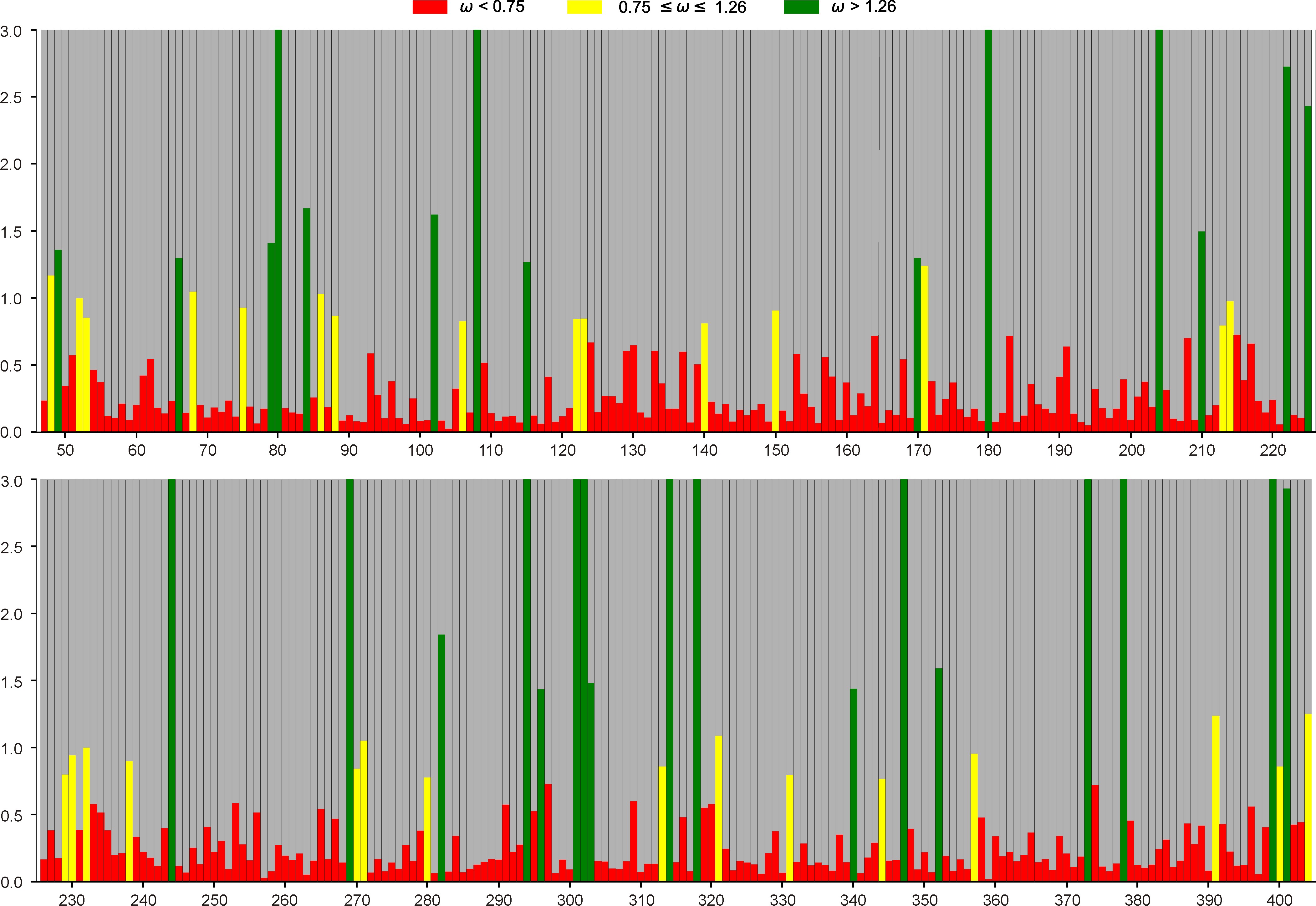
Figure 2. Result from the SIMMAP analysis showing that most of the amino acid positions are under negative selection (red) meaning that the dn/dn ratio was bellow 1.
Molecular homoplasies
We found molecular homoplasies within litostomatean alpha-tubulin. These homoplasies may cause nonsensical taxonomic branching in our alpha-tubulin trees. We hypothesize that these homoplastic positions arise by parallel evolution.
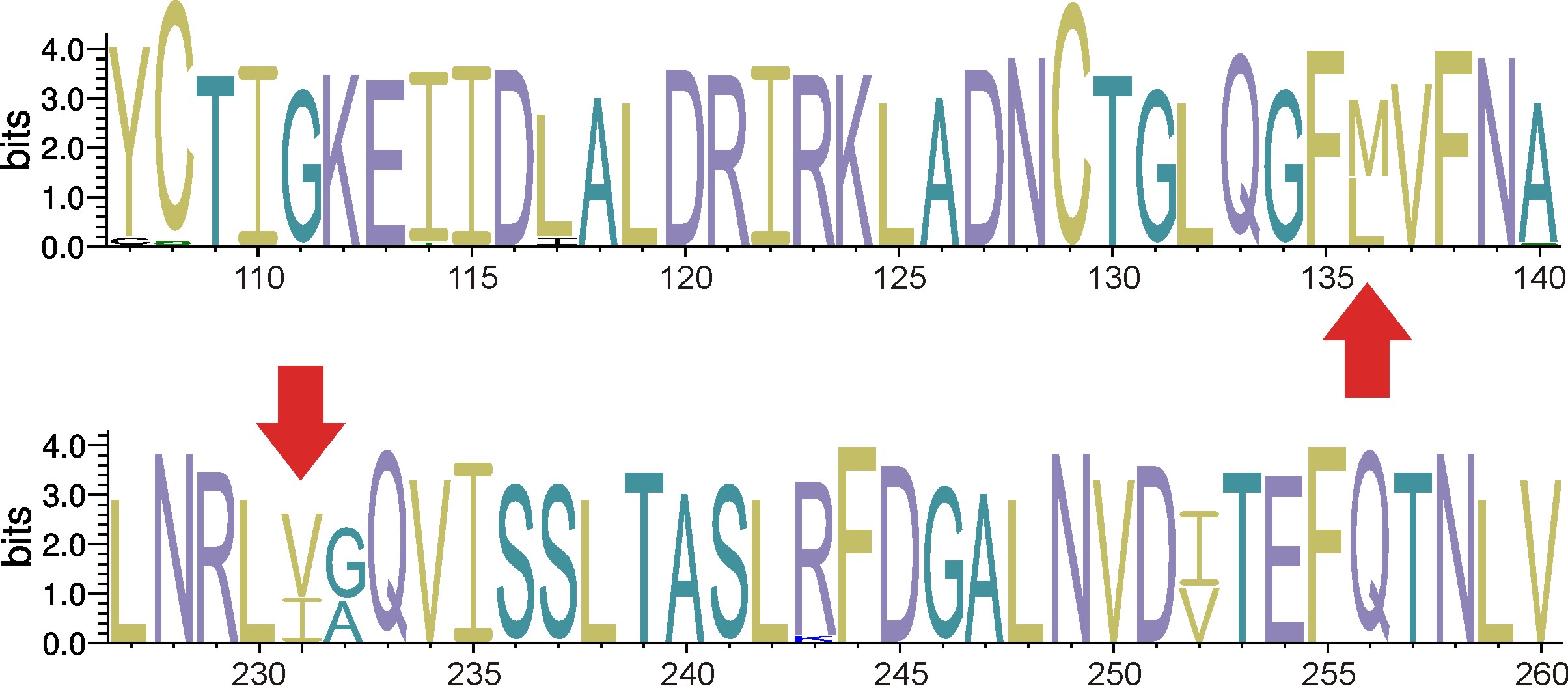
Figure 3. Section of the alpha-tubulin structure logo with the two homoplastic positions (arrows).
Conclusion
Our study showed that one should examine the information content and neutral evolution of protein-coding genes since different selective pressures may generate phylogenetic signal different from ancestry. Because the ciliate alpha-tubulin may be strongly affected by selection, great caution should be taken when tubulin genes are included in phylogenetic and/or phylogenomic analyses.